List of OGGM functions and tasks#
This page lists all available functions and classes in OGGM. It is a hard work to keep everything up-to-date, so don’t hesitate to let us know (see Get in touch) if something’s missing, or help us (see Contributing to OGGM) to write a better documentation!
Workflow#
Tools to set-up and run OGGM.
Read the configuration file containing the run's parameters. |
|
Same as initialise() but without requiring any download of data. |
|
Set the global logger parameters. |
|
Set the glacier intersection database for OGGM to use. |
|
Deletes the content of the working directory. |
|
Add an entry to the list of BASENAMES. |
|
Initializes the list of Glacier Directories for this run. |
|
Execute a task on gdirs. |
|
Run all flowline preprocessing tasks on a list of glaciers. |
|
Run all ice thickness inversion tasks on a list of glaciers. |
|
Shortcut function: run all tasks to merge tributaries to a main glacier |
|
Fit the total volume of the glaciers to the 2019 consensus estimate. |
Troubleshooting#
Prints the OGGM version and other system information. |
Input/Output#
Returns the path to the desired OGGM-sample-file. |
|
Path to the RGI directory. |
|
Path to the RGI region file. |
|
Get a list of glacier outlines selected from their RGI IDs. |
|
Path to the RGI directory containing the intersect files. |
|
Path to the RGI regional intersect file. |
|
Get a list of glacier intersects selected from their RGI IDs. |
|
Get the list of glaciers we have valid mass balance measurements for. |
|
Copies the glacier directories and their content to a new location. |
|
Writes the content of a glacier directory to a tar file. |
|
Merge the directories into 1000 bundles as tar files. |
|
Convert a glacier inventory into a dataset looking like the RGI (OGGM ready). |
|
Write the centerlines to a shapefile. |
|
Gather as much statistics as possible about a list of glaciers. |
|
Compiles the output of the model runs of several gdirs into one file. |
|
Merge the climate input files in the glacier directories into one file. |
|
Gathers the log output for the selected task(s) |
|
Gathers the time needed for the selected task(s) to run |
|
Compiles a table of specific mass balance timeseries for all glaciers. |
|
Gather as much statistics as possible about a list of glaciers. |
|
Compiles a table of ELA timeseries for all glaciers for a given years, using the mb_model_class (default MonthlyTIModel). |
OGGM Shop#
Returns a path to the desired CRU baseline climate file. |
|
Processes and writes the CRU baseline climate data for this glacier. |
|
Create a simple baseline climate file for this glacier - for testing! |
|
Returns a path to the desired ECMWF baseline climate file. |
|
Processes and writes the ECMWF baseline climate data for this glacier. |
|
Returns a path to the desired HISTALP baseline climate file. |
|
Processes and writes the HISTALP baseline climate data for this glacier. |
|
Applies the anomaly method to GCM climate data |
|
Processes and writes CESM climate data for this glacier. |
|
Read, process and store the CMIP5 and CMIP6 climate data for this glacier. |
|
Add the consensus thickness estimate to the gridded_data file. |
|
Reproject the its_live files to the given glacier directory. |
|
Initialize a glacier directory from an RGI-TOPO DEM. |
|
Select a DEM from the ones available in an RGI-TOPO glacier directory. |
|
Run a simple quality check on the rgitopo DEMs |
Entity tasks#
Entity tasks are tasks which are applied on single glaciers individually and do not require information from other glaciers (this encompasses the majority of OGGM’s tasks). They are parallelizable.
Very first task after initialization: define the glacier's local grid. |
|
Reads the DEM from the tiff, attempts to fill voids and apply smooth. |
|
Makes a gridded mask of the glacier outlines that can be used by OGGM. |
|
Compute glacier masks based on much simpler rules than OGGM's default. |
|
Writes a 1-0 glacier mask GeoTiff with the same dimensions as dem.tif |
|
Adds attributes to the gridded file, useful for thickness interpolation. |
|
Adds mass balance related attributes to the gridded data file. |
|
Writes a NetCDF variable to a georeferenced geotiff file. |
|
Compute the centerlines following Kienholz et al., (2014). |
|
Computes the Flowline along the unglaciated downstream topography |
|
The bedshape obtained by fitting a parabola to the line's normals. |
|
Compute the catchment areas of each tributary line. |
|
Computes the intersections between the catchments. |
|
Computes more physical Inversion Flowlines from geometrical Centerlines |
|
Compute geometrical catchment widths for each point of the flowlines. |
|
Corrects for NaNs and inconsistencies in the geometrical widths. |
|
Sets a new value for the terminus width. |
|
Compute "squeezed" or "collapsed" glacier flowlines from Huss 2012. |
|
Converts the "collapsed" flowline into a regular "inversion flowline". |
|
Adds the selected climate data to this glacier directory. |
|
Processes and writes the climate data from a user-defined climate file. |
|
Determine the mass balance parameters from a scalar mass-balance value. |
|
Calibrate for geodetic MB data from Hugonnet et al., 2021. |
|
Calibrate for in-situ, annual MB. |
|
Compute apparent mb from a linear mass balance assumption (for testing). |
|
Compute apparent mb from an arbitrary mass balance profile. |
|
Computes the mass balance with climate input from e.g. |
|
Computes the ELA of a glacier for a for given years and climate. |
|
Processes and writes the CRU baseline climate data for this glacier. |
|
Create a simple baseline climate file for this glacier - for testing! |
|
Processes and writes the HISTALP baseline climate data for this glacier. |
|
Processes and writes the ECMWF baseline climate data for this glacier. |
|
Applies the anomaly method to GCM climate data |
|
Processes and writes CESM climate data for this glacier. |
|
Read, process and store the CMIP5 and CMIP6 climate data for this glacier. |
|
Applies the anomaly method to historical climate data. |
|
Prepares the data needed for the inversion. |
|
Compute the glacier thickness along the flowlines |
|
Filters the last few grid points after the physically-based inversion. |
|
Small utility task to get to the volume od all glaciers. |
|
Surface velocities along the flowlines from inverted ice thickness. |
|
Compute a thickness map by redistributing mass along altitudinal bands. |
|
Compute a thickness map by interpolating between centerlines and border. |
|
Optimized search for a calving flux compatible with the bed inversion. |
|
Merges data from preprocessing tasks. |
|
Runs a model simulation with the default time stepping scheme. |
|
Runs the random mass balance model for a given number of years. |
|
Runs a glacier with climate input from e.g. |
|
Runs the constant mass balance model for a given number of years. |
|
Merges the output of two model_diagnostics files into one. |
|
Dynamically spinup the glacier to match area or volume at the RGI date. |
|
Calibrate melt_f to match a geodetic mass balance incorporating a dynamic model run. |
|
Copies the glacier directories and their content to a new location. |
|
Writes the content of a glacier directory to a tar file. |
Global tasks#
Global tasks are tasks which are run on a set of glaciers (most often: all glaciers in the current run). They are not parallelizable at the user level but might use multiprocessing internally.
Run all flowline preprocessing tasks on a list of glaciers. |
|
Run all ice thickness inversion tasks on a list of glaciers. |
|
Fit the total volume of the glaciers to the 2019 consensus estimate. |
|
Shortcut function: run all tasks to merge tributaries to a main glacier |
|
Get the list of glaciers we have valid mass balance measurements for. |
|
Write the centerlines to a shapefile. |
|
Compiles the output of the model runs of several gdirs into one file. |
|
Merge the climate input files in the glacier directories into one file. |
|
Gathers the log output for the selected task(s) |
|
Gathers the time needed for the selected task(s) to run |
|
Gather as much statistics as possible about a list of glaciers. |
|
Compiles a table of specific mass balance timeseries for all glaciers. |
|
Gather as much statistics as possible about a list of glaciers. |
|
Compiles a table of ELA timeseries for all glaciers for a given years, using the mb_model_class (default MonthlyTIModel). |
Command line interface (CLI)#
These commands are available:
oggm_netrc_credentialsoggm_preprooggm_benchmark
Generate the preprocessed OGGM glacier directories for this OGGM version |
Classes#
Listed here are the classes which are relevant at the API level (i.e. classes which are used and re-used across modules and tasks).
Organizes read and write access to the glacier's files. |
|
Geometry (line and widths) and flow rooting properties, but no thickness |
|
A Centerline with additional properties: input to the FlowlineModel |
|
Interface and common logic for all mass balance models used in OGGM. |
|
Monthly mass balance at given altitude(s) for a moment in time. |
|
Like self.get_monthly_mb(), but for annual MB. |
|
Specific mb for this year and a specific glacier geometry. |
|
Compute the equilibrium line altitude for a given year. |
|
Interface to OGGM's flowline models |
|
Duck FlowlineModel which actually reads data out of a nc file. |
Glacier directories#
Glacier directories (see also: GlacierDirectory) are folders
on disk which store the input and output data for a single glacier during
an OGGM run. The data are on disk to be persistent, i.e. they won’t be deleted
unless you ask OGGM to. You can start a run from an existing directory,
avoiding to re-do unnecessary computations.
Initialising a glacier directory#
The easiest way to initialize a glacier directory is to start from a pre-processed state available on the OGGM servers:
In [1]: import os
In [2]: import oggm
In [3]: from oggm import cfg, workflow
In [4]: from oggm.utils import gettempdir
In [5]: cfg.initialize() # always initialize before an OGGM task
# The working directory is where OGGM will store the run's data
In [6]: cfg.PATHS['working_dir'] = os.path.join(gettempdir(), 'Docs_GlacierDir')
# The base url is where to find the pre-processed directories
In [7]: base_url = 'https://cluster.klima.uni-bremen.de/~oggm/gdirs/oggm_v1.6/L1-L2_files/elev_bands/'
In [8]: gdirs = workflow.init_glacier_directories('RGI60-11.00897',
...: from_prepro_level=1,
...: prepro_base_url=base_url,
...: prepro_border=10)
...:
In [9]: gdir = gdirs[0] # init_glacier_directories always returns a list
We just downloaded the minimal input for a glacier directory. The
GlacierDirectory object contains the glacier metadata:
In [10]: gdir
Out[10]:
<oggm.GlacierDirectory>
RGI id: RGI60-11.00897
Region: 11: Central Europe
Subregion: 11-01: Alps
Name: Hintereisferner
Glacier type: Glacier
Terminus type: Land-terminating
Status: Glacier or ice cap
Area: 8.036 km2
Lon, Lat: (10.7584, 46.8003)
Grid (nx, ny): (139, 94)
Grid (dx, dy): (50.0, -50.0)
In [11]: gdir.rgi_id
Out[11]: 'RGI60-11.00897'
It also points to a specific directory in disk, where the files are written to:
In [12]: gdir.dir
Out[12]: '/tmp/OGGM/Docs_GlacierDir/per_glacier/RGI60-11/RGI60-11.00/RGI60-11.00897'
In [13]: os.listdir(gdir.dir)
Out[13]:
['diagnostics.json',
'outlines.tar.gz',
'dem.tif',
'glacier_grid.json',
'dem_source.txt',
'intersects.tar.gz',
'log.txt']
Users usually don’t have to care about where the data is located.
GlacierDirectory objects help you to get this information:
In [14]: fdem = gdir.get_filepath('dem')
In [15]: fdem
Out[15]: '/tmp/OGGM/Docs_GlacierDir/per_glacier/RGI60-11/RGI60-11.00/RGI60-11.00897/dem.tif'
In [16]: import rioxarray as rioxr
In [17]: rioxr.open_rasterio(fdem).plot(cmap='terrain');
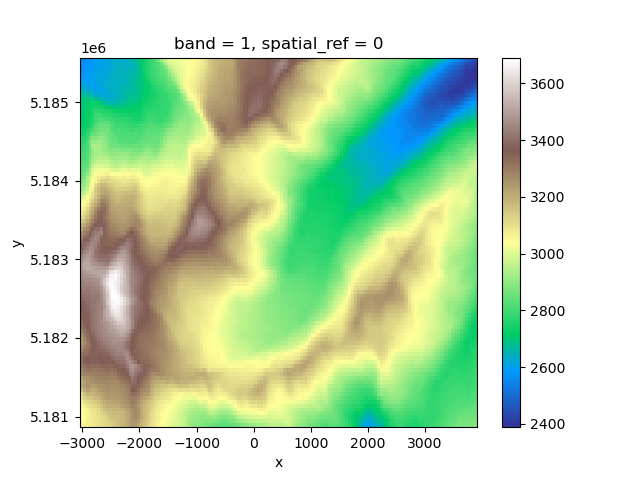
This persistence on disk allows for example to continue a workflow that has been previously interrupted. Initialising a GlacierDirectory from a non-empty folder won’t erase its content:
In [18]: gdir = workflow.init_glacier_directories('RGI60-11.00897')[0]
In [19]: os.listdir(gdir.dir) # the directory still contains the data
Out[19]:
['diagnostics.json',
'outlines.tar.gz',
'dem.tif',
'glacier_grid.json',
'dem_source.txt',
'intersects.tar.gz',
'log.txt']
For more information about how to use GlacierDirectories, visit our tutorial on the topic.
cfg.BASENAMES#
This is a list of the files that can be found in the glacier directory or
its divides. These data files and their names are standardized and listed in the
oggm.cfg module. If you want to implement your own task you’ll
have to add an entry to this file too.
- catchments_intersects.shp
The intersections between catchments (shapefile) in the local map projection.
- centerlines.pkl
A list of
oggm.Centerlineinstances, sorted by flow order.- climate_historical.nc
The historical monthly climate timeseries stored in a netCDF file.
- climate_historical_daily.nc
The historical daily climate timeseries stored in a netCDF file.(only temperature is really changing on daily basis,precipitation is just assumed constant for every day
- dem.tif
A geotiff file containing the DEM (reprojected into the local grid).This DEM is not smoothed or gap files, and is the closest to the original DEM source.
- dem_source.txt
A text file with the source of the topo file (GIMP, SRTM, …).
- diagnostics.json
A dictionary containing runtime diagnostics useful for debugging or logging of run parameters.
- downstream_line.pkl
A dictionary containing the downstream line geometry as well as the bed shape computed from a parabolic fit.
- elevation_band_flowline.csv
A table containing the Huss&Farinotti 2012 squeezed flowlines.
- fl_diagnostics.nc
A group netcdf file containing the model diagnostics (volume, thickness, velocity…) along the flowlines (thus much heavier).Netcdf groups = fl_{i}, with i between 0 and n_flowlines - 1
- flowline_catchments.shp
Each flowline has a catchment area computed from flow routing algorithms: this shapefile stores the catchment outlines (in the local map projection).
- gcm_data.nc
The monthly GCM climate timeseries stored in a netCDF file.
- geometries.pkl
A dictionary containing the shapely.Polygons of a glacier. The “polygon_hr” entry contains the geometry transformed to the local grid in (i, j) coordinates, while the “polygon_pix” entry contains the geometries transformed into the coarse grid (the i, j elements are integers). The “polygon_area” entry contains the area of the polygon as computed by Shapely. The “catchment_indices” entrycontains a list of len n_centerlines, each element containing a numpy array of the indices in the glacier grid which represent the centerlines catchment area.
- glacier_grid.json
A
salem.Gridhandling the georeferencing of the local grid.- glacier_mask.tif
A glacier mask geotiff file with the same extend and projection as the dem.tif. This geotiff has value 1 at glaciated grid points and value 0 at unglaciated points.
- gridded_data.nc
A netcdf file containing several gridded data variables such as topography, the glacier masks, the interpolated 2D glacier bed, and more. This is for static, non time-dependant data.
- gridded_simulation.nc
A netcdf file containing gridded data variables which are time dependant. It has the same coordinates as gridded_data.
- hypsometry.csv
A hypsometry file computed by OGGM and provided in the same format as the RGI (useful for diagnostics).
- intersects.shp
The glacier intersects in the local map projection.
- inversion_flowlines.pkl
A “better” version of the centerlines, now on a regular spacing i.e., not on the gridded (i, j) indices. The tails of the tributaries are cut out to make more realistic junctions. They are now “1.5D” i.e., with a width.
- inversion_input.pkl
List of dicts containing the data needed for the inversion.
- inversion_output.pkl
List of dicts containing the output data from the inversion.
- linear_mb_params.pkl
When using a linear mass balance for the inversion, this dict stores the optimal ela_h and grad.
- mb_calib.json
A dict containing the glacier’s mass balance calibration parameters.
- model_diagnostics.nc
A netcdf file containing the model diagnostics (volume, mass balance, length…).
- model_flowlines.pkl
List of flowlines ready to be run by the model.
- model_geometry.nc
A netcdf file containing enough information to reconstruct the entire flowline glacier geometry along the run (can be expensive in disk space).
- outlines.shp
The glacier outlines in the local map projection (Transverse Mercator or UTM).
Mass balance#
Mass balance models found in the core.massbalance module.
They follow the MassBalanceModel interface. Here is a quick summary
of the units and conventions used by all models:
Units#
The computed mass balance is in units of [m ice s-1] (“meters of ice per
second”), unless otherwise specified (e.g. for the utility function
get_specific_mb).
The conversion from the climatic mass balance ([kg m-2 s-1] ) therefore assumes
an ice density given by cfg.PARAMS['ice_density'] (currently: 900 kg m-3).
Time#
The time system used by OGGM is a simple “fraction of year” system, where the floating year can be used for conversion to months and years:
In [20]: from oggm.utils import floatyear_to_date, date_to_floatyear
In [21]: date_to_floatyear(1982, 12)
Out[21]: 1982.9166666666667
In [22]: floatyear_to_date(1.2)
Out[22]: (1, 3)
Interface#
Interface and common logic for all mass balance models used in OGGM. |
|
Monthly mass balance at given altitude(s) for a moment in time. |
|
Like self.get_monthly_mb(), but for annual MB. |
|
Specific mb for this year and a specific glacier geometry. |
|
Compute the equilibrium line altitude for a given year. |
Models#
Constant mass balance as a linear function of altitude. |
|
Monthly temperature index model. |
|
Constant mass balance during a chosen period. |
|
Random shuffle of all MB years within a given time period. |
|
Adding uncertainty to a mass balance model. |
|
Handle mass balance at the glacier level instead of flowline level. |
Model Flowlines#
Flowlines are what represent a glacier in OGGM. During the preprocessing stage of a glacier simulation these lines evolve from simple topography following lines to more abstract objects within the model.
This chapter provides an overview on the different line types, how to access the model flowlines and their attributes.
Hierarchy#
First a short summary of the evolution or the hierarchy of these different flowline stages:
- geometrical centerlines
These lines are calculated following the algorithm of Kienholz et al., (2014). They determine where the subsequent flowlines are situated and how many of them are needed for one glacier. The main task for this calculation is
compute_centerlines(). These lines are not stored within a specific OGGM class but are stored within the GlacierDirectory as shapely objects.- inversion flowlines
To use the geometrical lines within the model they are transformed to
Centerlineobjects. This is done withininitialize_flowlines()where the lines are projected onto a xy-grid with regular spaced line-points. This step also takes care of tributary junctions. These lines are then in a later step also used for the bed inversion, hence their name.- downstream line
This line extends the glacier’s centerline along the unglaciated glacier bed which is necessary for advancing glaciers. This line is calculated along the valley floor to the end of the domain boundary within
compute_downstream_line().
- model flowlines
The dynamical OGGM model finally uses lines of the class
Flowlinefor all simulations. These flowlines are created byinit_present_time_glacier(), which combines information from several preprocessing steps, the downstream line and the bed inversion. From a user perspective, especially if preprocessed directories are used, these model flowlines are the most important ones and further information on the class interface and attributes are given below.
Access#
The easiest and most common way to access the model flowlines of a glacier is with its GlacierDirectory. For this we initialize a minimal working example including the model flowlines. This is achieved by choosing preprocessing level 5.
In [23]: import os
In [24]: import oggm
In [25]: from oggm import cfg, workflow, DEFAULT_BASE_URL
In [26]: from oggm.utils import gettempdir
In [27]: cfg.initialize() # always initialize before an OGGM task
# The working directory is where OGGM will store the run's data
In [28]: cfg.PATHS['working_dir'] = os.path.join(gettempdir(), 'Docs_GlacierDir2')
# The prepro_base_url is where to find the pre-processed directories
In [29]: gdirs = workflow.init_glacier_directories('RGI60-11.00897',
....: from_prepro_level=5,
....: prepro_base_url=DEFAULT_BASE_URL,
....: prepro_border=80)
....:
In [30]: gdir = gdirs[0] # init_glacier_directories always returns a list
In [31]: fls = gdir.read_pickle('model_flowlines')
In [32]: fls
Out[32]: [<oggm.core.flowline.MixedBedFlowline at 0x7f666df260d0>]
In [33]: [fl.order for fl in fls]
Out[33]: [0]
This glacier has three flowlines of type MixedBedFlowline provided as a list. And the flowlines are ordered by ascending Strahler numbers, where the last entry in the list is always the longest and very often most relevant flowline of that glacier.
Type of model flowlines#
A Centerline with additional properties: input to the FlowlineModel |
|
A Flowline which can take a combination of different shapes (default) |
|
A parabolic shaped Flowline with one degree of freedom |
|
Simple shaped Flowline, glacier width does not change with ice thickness |
|
A Flowline with trapezoidal shape and two degrees of freedom |
Important flowline functions#
Computes more physical Inversion Flowlines from geometrical Centerlines |
|
Compute the centerlines following Kienholz et al., (2014). |
|
Computes the Flowline along the unglaciated downstream topography |
|
Merges data from preprocessing tasks. |
